Liu Yu’s team Nat Chem | at Shanghai Jiao Tong University used engineered DNA polymerase variants to introduce multiple modifications at specific sites in RNA
Using PORDVA innovative technology, fine “pack” RNA
There is a growing need for precise position modification of large Rnas in biomedical research and therapeutic applications.
These modifications are critical to understanding the function of RNA, developing new diagnostic tools, and creating more effective treatments.
However, existing RNA synthesis methods are either inefficient or unable to be adapted to RNA molecules beyond a certain length, which limits scientists’ ability to conduct in-depth research and innovative applications of RNA.
In addition, some methods rely on special chemical reagents, which makes it difficult for researchers who do not have chemical synthesis platforms to take advantage of these techniques.
Therefore, the development of an efficient, universal and suitable for various RNA modification methods has become an important goal in the field of scientific research.
PORDVA (position-specific labelling of RNA by DNAP variant) technology is an innovative RNA modification method that utilizes engineered DNA polymerase (DNAP) variants to introduce multiple modifications at specific locations in RNA.
Through carefully designed DNAP variants, single-stranded DNA templates, and RNA strands that serve as primers, PORDVA is able to precisely add a variety of chemical groups including base modification, 2′ -ribose modification, and skeleton modification during RNA synthesis.
This technique not only achieves a modification efficiency of more than 85%, but is also able to process RNA molecules up to thousands of nucleotides, greatly expanding the boundaries of RNA research and application.
The successful development of PORDVA technology provides an efficient, versatile and flexible tool for the functionalization of RNA, which is expected to lead to changes in many fields such as basic research, biotechnology and medical therapy.
Yu Liu’s team successfully introduced a variety of modifications, including fluorophore, natural modifications and biotin, on a variety of RNA molecules and demonstrated the effects of these modifications on RNA stability and protein production, providing a powerful new tool for basic research and therapeutic applications of RNA.
They published in Nature Chemistry under the title “Engineering a DNA polymerase for modifying large RNA at specific positions.” It is the first paper of Nature Chemistry published by Shanghai Jiao Tong University this year.
【 Main content 】
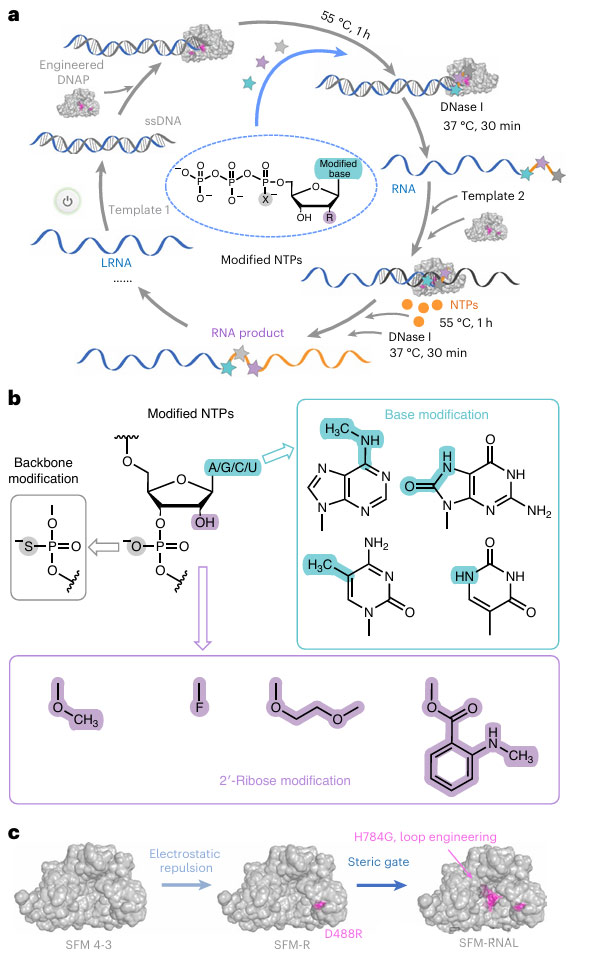
The synthesis of PORDVA is depicted in detail, including an Engineered DNA polymerase (Engineered DNAP), a single-stranded DNA template (ssDNA), and a strand of RNA (LRNA) that acts as a primer.
LRNA pairs complementally with the 3′ end of the ssDNA template, and engineered DNA polymerase uses labeled nucleotide triphosphate (NTPs) for RNA extension, thereby introducing modifications at specific sites.
The modified RNA can released by DNase I after digesting the DNA template, either as a final product or as LRNA for subsequent PORDVA reactions.
The figure also shows the types of modifications that can introduced into RNA, including base modifications, skeleton modifications, and 2′ -ribose modifications, which precisely integrated into RNA using PORDVA technology.
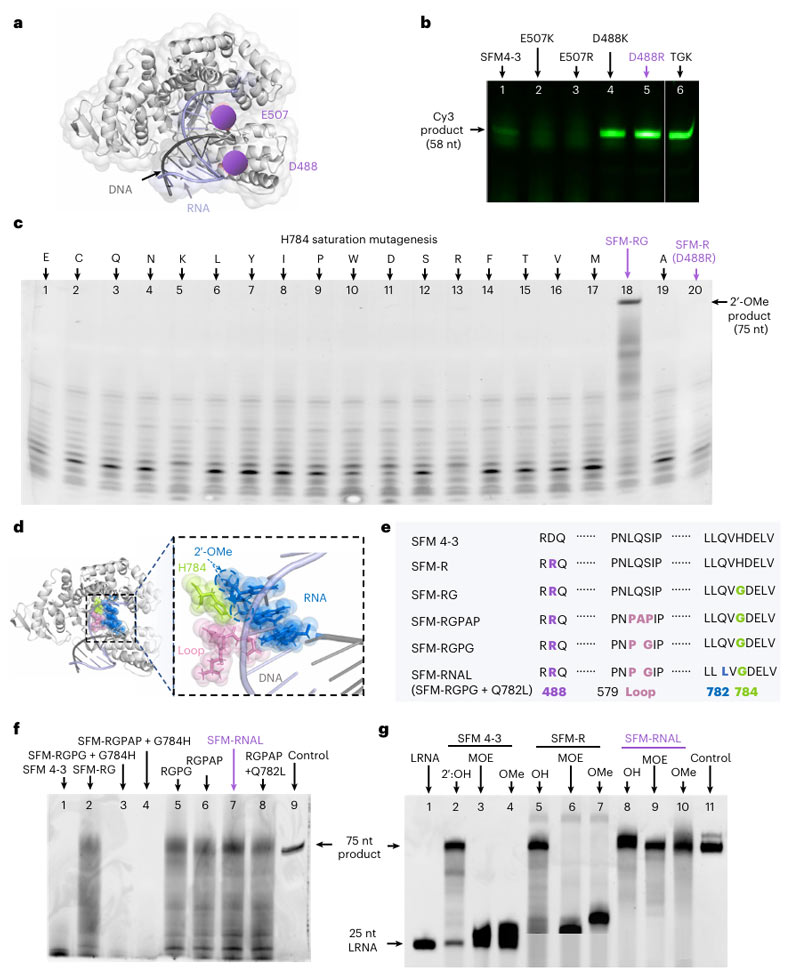
First, the crystal structure of Taq DNAP is presented, with two acidic amino acid residues E507 and D488, which interact with nucleic acid double-stranded, specifically labeled.
By mutating these two residues into basic amino acids, such as E507K/R and D488K/R, the researchers aim to reduce electrostatic repulsion between DNAP and the nucleic acid double strand, thereby increasing the efficiency of RNA elongation.
The experimental results showed that the D488R mutant (also known as SRM-R) showed a significant efficiency improvement in the Cy3-labeled PORDVA response.
Further optimization of SFM-R through site-saturation mutation and ring engineering to improve its tolerance to 2′ -O-methyl (2′-OMe) nucleotides also demonstrated, resulting in the identification of SFM-RG (D488R and H784G) mutants capable of efficient synthesis of full-length 2′ -OME-RNA.
These findings provide an important structural basis and optimization strategy for the application of PORDVA technology in RNA modification.
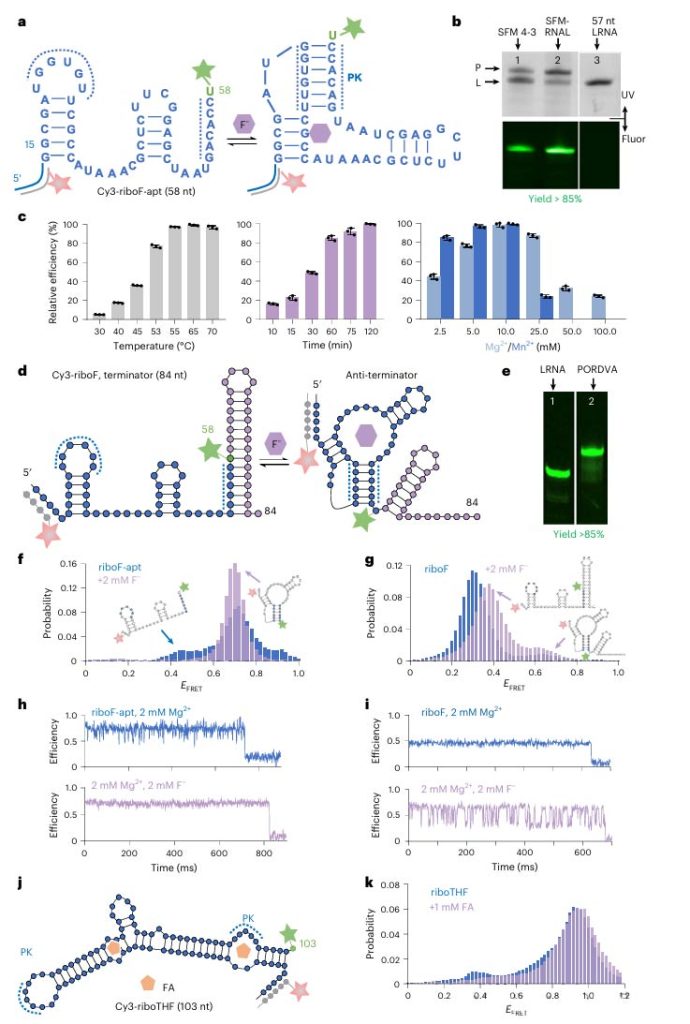
Using PORDVA technology, the researchers successfully introduced Cy3 fluorophore at a specific location in the riboF, achieving a modification efficiency of up to 85% by optimizing reaction conditions, including temperature, time, and bivalent cation concentration.
The results of single molecule fluorescence resonance energy transfer (smFRET) experiments show that the modified riboF exhibits different properties in structure from the unmodified riboF, especially in the presence of fluoride ions, and the modified riboF is more inclined to form a high FRET conformation containing pseudo-junctions (PK), while the unmodified riboF rarely forms such a conformation.
These results not only validate the effectiveness and accuracy of PORDVA technology in RNA modification, but also provide a new perspective for understanding the structural changes and functional regulation of RNA molecules.
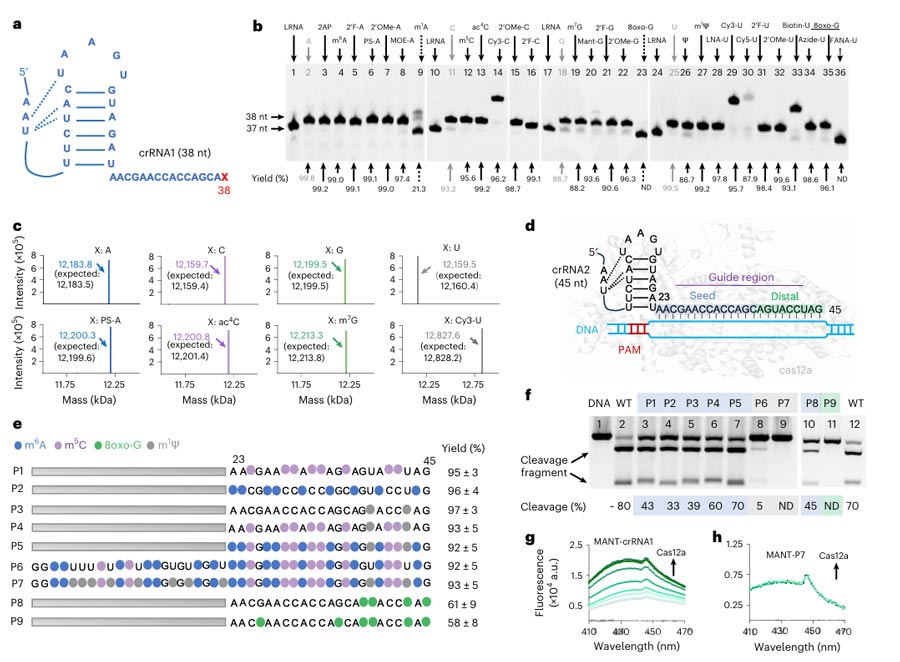
The researchers used PORDVA technology to introduce more than 20 different modifications at specific locations in crRNA, including fluorophores, natural modifications, and chemical groups.
Using gel electrophoresis and liquid chromatography mass spectrometry (LC-MS) analysis, the researchers verified the efficient introduction of these modifications and observed the effects of these modifications on CRISPR-Cas12a function.
For example, some modifications significantly reduced the efficiency of CRISPR-Cas12a’s DNA cutting, while others had less impact on function.
These results indicate that PORDVA technology can not only efficiently introduce multiple modifications on crRNA, but also fine-regulate the gene editing function of CRISPR-Cas12a through these modifications, providing a new strategy for the development of more efficient and more specific gene editing tools.
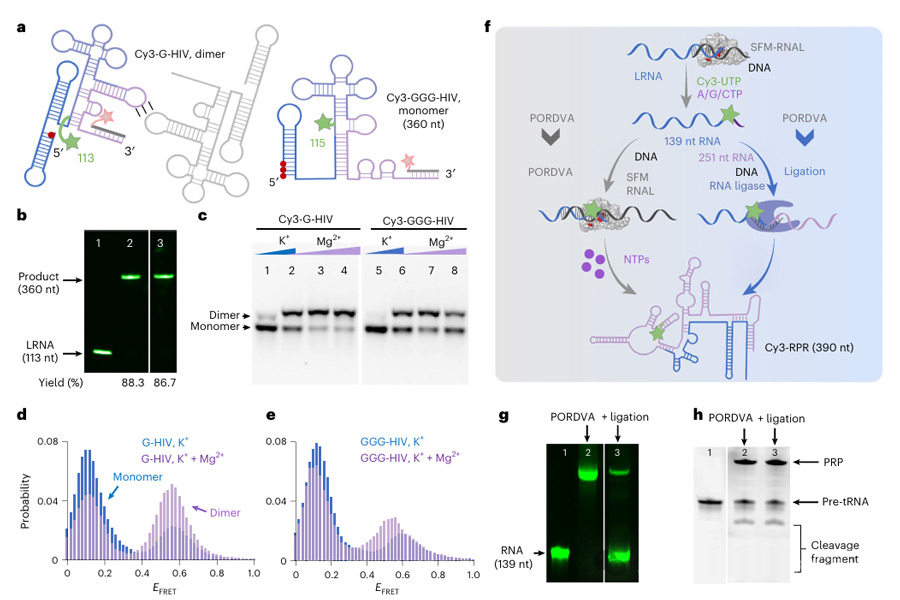
The researchers used PORDVA technology to introduce Cy3 fluorophores at specific locations of HIV RNA, and observed the dimer formation of HIV RNA with different structures under physiological conditions through single-molecule Forster resonance energy transfer (smFRET) experiments.
In addition, the researchers also fluorescently labeled the internal position of E. coli RNase P RNA, successfully labeled RPR by PORDVA alone or in combination with the ligand reaction, and verified the activity of labeled RPR in catalyzing the maturation of the precursor tRNA.
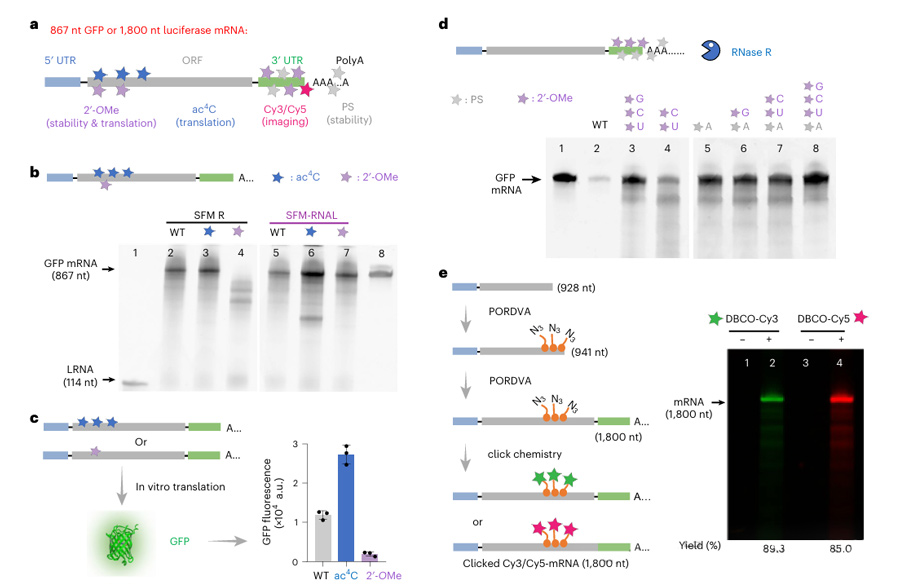
We used PORDVA technology to introduce modifications including N4-acetylcytosine (ac4C) and 2′ -O-methyl (2′-OMe) in the open reading frame (ORF) of GFP mRNA, and 2′-OMe and phosphothioate (PS) modifications in the 3′ -untranslated region (3′-UTR) and the tail of polyA.
These modifications significantly improved mRNA stability and translation efficiency.
For example, AC4C-modified mRNA showed a significant improvement in translation efficiency, while 2′-OMe-A modified inhibited the translation of GFP mRNA.
In addition, the researchers introduced azide groups in the 3′-UTR and ORF regions of the mRNA, which further modified to Cy5 or Cy3 labeled mRNA by a click chemical reaction.
These results indicate that PORDVA technology can not only efficiently introduce multiple modifications on mRNA, but also fine-regulate mRNA stability and translation efficiency through these modifications, providing a new strategy for the development of more stable and effective mRNA drugs and vaccines.
【 Summary and Prospect 】
This paper describes an innovative technique called PORDVA, which uses engineered DNA polymerase variants to introduce multiple modifications at specific locations in RNA.
Through semi-rational design, the researchers successfully developed highly efficient DNA polymerase variants that can introduce a variety of chemical groups, including base modification, 2′ -ribose modification, and skeleton modification, at specific sites of RNA, with an efficiency of more than 85%.
PORDVA technology is not only applicable to a variety of RNA molecules, such as fluororibose switch (riboF), CRISPR-cas12 guide RNA (gRNA), HIV RNA, ribonuclinase P (RNase P) and messenger RNA (mRNA).
RNA structural and functional changes can also be studied using techniques such as single-molecule Forster Resonance Energy Transfer (smFRET).
In addition, PORDVA technology performs well in the modification of mRNA, can significantly improve the stability and translation efficiency of mRNA, and provides a new strategy for the development of more stable and effective mRNA drugs and vaccines.
Looking forward to the future, PORDVA technology expected to trigger changes in many fields such as basic research of RNA, biotechnology and medical therapy, and promote the further development of RNA-related research and applications.